Cell Energy
---------------------
created 7/07
updated 1/30/13
My related essay on Photosynthesis is here: Photosynthesis
Go to homepage
Here is an
essay on how eukaryotic (animal and plant) cells use energy (from imported
high energy glucose molecules) to control their ion concentrations, membrane
voltage, water, and do various specialized jobs.
-------------------------------------------------------------------------
Reference
1 ) 1 ev =
23.1 kcal/mol = 96.5 kj/mol
-------------------------------------------------------------------------
Introduction
Learning about
photosynthesis with its specialized membrane processes powered by photon
energy made me curious about how cells (in general) handle energy. Amazingly
no reference I have found gives the big picture on energy, it's all details.
As I have learned more about cells, there does appear to be a relatively
simple big picture about how from an energy viewpoint cells function.
Big, big picture -- cell energy and photosynthesis
The ways in which energy in a cell is used has a lot in common with the
ways plant chloroplasts capture the energy from solar photons. Consider
In a cell
* local regions in the cell (mitochondria) use energy from imported
glucose to make
a proton gradient
* energy from the proton gradient is transferred (by ATP synthase)
to (recycled)
ATP molecules
-- glucose (C6H12O6) has atoms of only carbon, oxygen and hydrogen. The
oxidation of glucose releases its hydrogen and oxygen as a water byproduct
and imported oxygen (from respiration) combines with the carbon of
glucose to form CO2 byproduct. The oxidization of glucose releases
2,870 kJ/mole of energy (which went into making it) about half of which
is transferred to the ATP molecules (50% conversion efficiency).
* energy extracted from ATP molecules powers the major ion pump
of the cell
(generates K+ and Na+ gradients), and runs many cellular functions, like
generating heat, powering muscles and flagella
* energy from the K+/Na+ gradient is used by many other membrane
pumps and
transporters
In a chloroplast
*
local regions in the chloroplast (photosystems) use electrons energized
by photons
to make a proton gradient
* energy from the proton gradient is transferred (by ATP synthase)
to (recycled)
ATP molecules
* energy extracted from ATP molecules run local cellular functions,
like ion pumps
to make ion gradients
* local regions of chloroplasts use energy from ATP to make glucose
which is exported
as an energy source to the rest of the plant
-- glucose (C6H12O6) has atoms of only carbon, oxygen and hydrogen. The
carbon and oxygen come from CO2 (pulled from the air) and the hydrogen
from water (pulled up by the roots). Excess oxygen (from the water) is
released into the air as a byproduct. Synthesis of glucose requires an
input
of 2,870 kJ/mole of energy, all of which comes from captured solar
photons.
Cell energy overview
All animal
and plant cells (eukaryotic cells) take in fuel in the form of sugar (glucose)
and bring it into microchondria, which are separate, isolated regions of
the cell (organelles), where it is burned (oxidized) producing by-products
CO2 and H2O. The microchondria use the energy obtained from the sugar to
make the energy rich molecule ATP that is exported into the cell.
ADP/ATP is
an energy carrier molecule within the cell. Energy is stored in the molecule
by attaching (via a covalent bond) an extra phosphate (phosphorus atom
with oxygens), and energy is extracted by popping off a phosphate. It cycles
between having two phosphate groups (ADP is Adenosine Di-Phosphate)
and three phosphate groups (Adenosine Tri-Phosphate). So microchondria
take in ADP and add energy by adding a phosphate group creating ATP. Various
processes in the cell, like ion pumps in the membrane, get the energy they
need by popping of a phosphate group from ATP, taking it back to ADP.
The cell uses
a lot of the ATP stored energy to run ion pumps. The cell pumps in
potassium,
and pumps out salt (sodium and chloride) and calcium. The pumps
regulate the concentration of the major ions in the cell fluid, keeping
potassium ion concentration always high and the sodium, calcium,
and chloride ion concentration always low. The big, main pump of
the cell is the Na+/K+ ATPase transporter, which pumps potassium
(in) and sodium (out), other pumps handle calcium and chloride ions. The
pumps are large complex proteins that span the cell membrane.
The enzymes
that use ATP for energy (ATPases) perform what the chemist call a coupled
reaction.
Some of the energy released when the phosphate bond is broken (exothermic
reaction) is captured to perform work and the rest goes into heat. One
'turn' of the Na+/K+ ATPase pump uses the energy of one ATP (ATP => ADP
+ Pi) to pump 3 Na+ out of the cell and 2 K+ in. (net charge flow
outward)
Most of the
energy that the pumps expend building up the concentration gradients is
not lost, it is stored in the concentration gradients that the pumps create.
This energy is available to other proteins that live in the membrane. By
opening an ion diffusion channel across the membrane energy can be extracted
from ions diffusing down their concentration gradient. One example of this
is the Na+/glucose pump. The energy it needs to pump glucose into
the cell comes from it providing a channel for Na+ ions to diffuse into
the cell, down their concentration gradient.
Another
(classic) example of using concentration gradients for energy occurs in
chloroplasts doing photosynthesis. Here a high gradient (10,00 to 1) of
protons (H+) pumped up by the energy from captured light photons is used
as a source of energy to make ATP in the chloroplast's membrane. This process
is called chemiosmosis (by analogy with osmosis) and the enzyme that makes
ATP is called ATP synthase. The understanding of this has been called
one of the seminal discoveries in biology in the 20th century and won Mitchell
Nobel prize for Chemistry in 1978.
Mitochondria
organelles in cells also make ATP using a proton gradient and the enzyme
ATP synthase. In this case the proton gradient is only about 10 to 1 and
the energy needed to create it comes from the oxidization of glucose. The
protons just circulate around, pumped in and then diffusing back out. The
potential across the mitochondria membrane is -200 mv (inside negative).
(Bacteria make ATP using chemiosmosis too.)
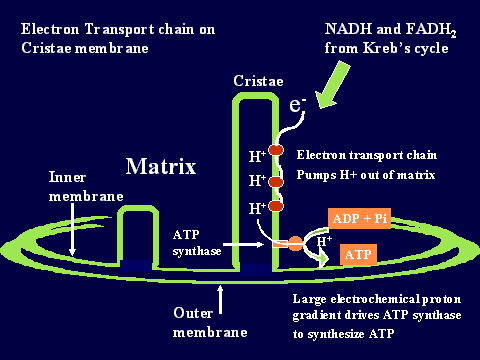
Synthesis of ATP by ATP synthase in mitochondria
In mitochondria the PMF (proton motive force) is almost entirely made up
of the electrical component but in chloroplasts the PMF is made up mostly
of the pH gradient. In either case the PMF needs to be about 50 kJ/mol
for the ATP synthase to be able to make ATP.
Human body ATP energy
Wikipedia
has the following interesting perspective on ATP in human cells:
The
energy used by human cells requires the hydrolysis of 50 to 75 kg of ATP
daily, which means a human typically will use up their body weight of ATP
over the course of the day! However, the total quantity of ATP in
the human body (at any one time) is about 50 mg. This means that each ATP
molecule is recycled 1,000 to 1,500 times during a single day (50 kg/50
mg = 1000). (1,000 times a day is each ATP being recycled every 86
sec). ATP cannot be stored, hence its consumption closely follows its synthesis.
For fun we can
calculate how much power is being processed by ATP in the human
body. The molecular weight of ATP is about 500, so 50 mg is about 0.1 mole,
and according to Widipedia (see above) this amount of ATP releases its
energy 1,000 times a day. Power (watt) =energy (joule)/time(sec).
ATP (combined with water)
30.5 kj/mole (7.3 kcal/mole)
Energy (0.1 mole ATP)
3.05 kj
Power (0.1 mole every 86 sec)
3,050 j/86 sec
35 watt
I found a reference
that states the efficiency of energy transfer from glucose to ATP (in mitochondria)
is approximately 50%. This means 70 watts of food energy are needed to
get 35 watts into ATP.
Check
If you eat 2,000 (nutritional) calories/day, this is (2,000 kcal/day x
4.18 j/cal) = 8.36 million j/day. A day has 86,400 sec, so the power
provided by eating 2,000 cal/day is about 100 watts. Hence, about 75% of
2,000 food calories is used in the ATP synthesis process with about half
of this energy making its way into ATP. Checks.
Another interesting,
related topic is how the body cools itself, How does it dump the 100 watts
or so that it is continually extracting from oxidizing food. There are
three major routes for heat to leave the body: convection, radiation, and
respiration. Here some estimates I found which look reasonable (for low
activity):
Convection (heat transfer from skin to air)
65 watt
Radiation (skin 6F warmer than ambient)
24 watt
Respiration (exhaled warmed air & H2O)
11 watt
------------------------
total 100 watt
Organelles
Organelles
in eukaryotic cells (cells of everything except bacteria and archaea) are
sort of like cells within cells. Wikipedia says, "an organelle is to the
cell what an organ is to the body." Organelles are small regions of the
cell, isolated from the rest of the cell with a membrane, that do specific
jobs for the cell, and even have their own DNA.
Examples of
organelles are microchondria, which oxidize fuels to make energy for the
cell and chloroplasts, which do photosynthesis. There can be a huge number
of organelles in a cell. A typical eukaryotic cell contains about 2,000
mitochondria (about 20% of its volume). A good case can be make that
organelles chloroplasts and microchondria were originally free living bacteria
(cynobacteria) that got captured (a process called endosymbiosis) by plant
and animal cells.
Cell membrane voltage (overview)
All cells
normally maintain a voltage across their cell membranes (-80mv or so).
They use this voltage to pull molecules they need (like sugars)
into the cell from the outside. The E field across the cell membrane points
inward, so the cell can only pull in (via this mechanism) positively charged
outside molecules or ions. Glucose (sugar) get pumped in with positive
sodium ions (Na+) though channels where energy is available from the sodium
ions that feel an inward force from both the E field and the sodium concentration
gradient.
Cells generate
the membrane potential by a two step process. First they do work ('burning'
fuel ATP) pumping ions (mostly potassium K+) into the cell. This
pumping does not (directly) produce a membrane potential, because (somehow)
the inside of the cell is initially neutral with a balancing negative charge
residing on proteins in the cell. (Are electrons being pumped in too?)
It's important to note that the positive charge resides on something tiny
and mobile (nucleus of an atom) compared to the carrier of the negative
charge (protein) that is large and relatively immobile.
Second step
is open (potassium) ion channels allow K+ ions (only) to begin to passively
diffuse down the concentration gradient toward the outside. K+ ions actually
diffuse through the ions channels in both directions, but because the concentration
of K+ inside the cell is much higher than outside the net result is diffusion
of positive ions out of the cell. The negative charge, unlike the positive
charge, is unable to get out of the cell, because it resides on proteins,
which are far too large to fit into the small, open, ion channels. The
(net) diffusion down these ion channels soon stops, because the net negative
charge inside the cell cause an inward pointing E field to appear that
opposes the outward diffusion pressure of the positive ions.
A slightly
more technical (& obscure) explanation I found puts it this way:
There
are two forces acting on a given ionic species. The driving force of the
chemical concentration gradient tends to move ions down this gradient (chemical
potential). On the other hand the electrostatic force due to the charge
separation across the membrane tends to move ions in a direction determined
by its particular charge. Eventually an equilibrium can be reached so that
the actual ratio of intracellular and extracellular concentration ultimately
depends on the existing membrane potential.
Element ions that are
positive, like potassium, sodium and calcium, live on the left side of
the periodic chart where electrons are easily lost. Elements like chlorine
that live on the right side of the periodic chart easily gain electrons,
so they become negative ions.
Ion transport across membranes
Cells normally
keep ion levels inside (relative to fluid outside) as shown below.
The second column lists the permeability (which looks like a diffusion
constant). These values are for mammalian cells.
ion
concentration
diffusion rate
----------------
------------------
-------------------
potassium (K+)
high (x 20)
5x 10^-7 cm/sec
chloride (Cl-)
low (1/10)
1x 10^-8 cm/sec
sodium (Na+)
low (1/10)
5x 10^-9 cm/sec
calcium (Ca2+)
very low (1/10,000)
Note cells
normally pump up their (internal) potassium concentration and pump
down
the
concentrations of all the other ions (sodium, chloride and calcium).
These ion flows
do not imply a (substantial) charge imbalance. I'm not sure how this is
managed but (apparently) electron (?) and proton concentrations are adjusted
too. A small charge imbalance, say due to a loss of positive internal charge,
is able to produce a (typical) -70mv internal voltage. All that is needed
is to open a potassium diffusion channel. After a small quantity of potassium
ions (positive charge) diffuse out a voltage and electric field then develops
across the membrane pointing inward, which pushes the positive ions inward
stopping the diffusion. (Well really what happens is that diffusion currents
outward are canceled by electric drift currents inward.)
Also note that
(rapid) flows down the concentration gradient of either (or both) of the
lower two ions (Na+ and Ca2+) can (rapidly) 'flip' the membrane voltage,
driving the inside of the cell positive, and that concentration gradient
flows of K+ and/or Cl- can reverse the flip. This is in fact how so called
'active cells' like nerve and muscle cells work, followed by recovery period
(refractory period) while the cell's ion pumps restore the concentration
gradients.
Na+/K+ ATPase transporter
Cells (eukaryote
cells of animals and plants) have ion pumps in their membrane (powered
by burning ATP) that pump potassium ions (K+) into the cell and
pump sodium ions (Na+) out of the cell. In fact the same pump does
both jobs (Na+/K+ ATPase transporter), and it changes concentration levels
about a factor of 10. References say for every two K+ pumped in, three
Na+ are pumped out. (Is this ratio fixed? Does this lead to a loss of charge
in the cell?) Below is a link to a neat animation that shows the
Na+/K+ ATPase transporter working (inside of cell is below).
http://www.stolaf.edu/people/giannini/flashanimat/transport/secondary%20active%20transport.swf
The crucial
roles of the Na+/K+ ATPase are reflected in the fact that almost one-third
of all the energy generated by the mitochondria in animal cells is used
just to run this pump. It looks (to me) that the Na+/K+ ATPase pump is
the primary engine of the cell. The potential energy that this (fuel
burning) pump stores up in the concentration gradients (of K+ and Na+)
and in the membrane electric field is available to be tapped by other (secondary)
engines that do work in the cell, for example, the Na+/glucose transporter
(below) that brings in glucose.
Mitochondria
are organelles in cells that make ATP (used for energy in the membrane
and throughout the cell) by oxidizing food molecules (glucose) breaking
them down into CO2 and water. The sugars are the external source of energy
to the cell. The 'eating' of sugars (food molecules) by mitochondria inside
cells is sometimes called (misleadingly) cellular respiration.
Na+/glucose transporter
One the right
side of the (above) animation some of the pumped out Na+ ions are shown
diffusing back in bringing in glucose molecules with them. In the long
term, of course, all ions pumped one way must later diffuse back (generally
through other paths) to keep the cell's ion concentration and charge in
steady state. Here is a more technical description of the Na+/glucose pathway:
Na+/glucose transporter --- This transmembrane
protein allows sodium ions and glucose to enter the cell together. The
sodium ions flow down their concentration gradient while the glucose molecules
are pumped up theirs. Later the sodium is pumped back out of the cell by
the Na+/K+ ATPase. Note an ion flowing down its concentration gradient
can do work. The energy needed to pump the glucose is being extracted from
the sodium, which is why their paired. Technically this pairing is called
'Indirect Active Transport'.
There is a similar
pump for calcium ions using the sodium concentration gradient. The Na+/Ca2+
exchanger pulls energy from Na+ diffusing into the cell to pump Ca2+ out
of the cell. Cells normally maintain low levels of calcium inside. There
is also an ATP burning removal pump for calcium called the Ca2+ ATPase
transporter. Here is Wikipedia describing the sodium/calcium pump:
"Na+/Ca2+ exchanger uses the energy that is stored
in the electrochemical gradient of sodium (Na+) by allowing Na+ to flow
down its gradient across the plasma membrane in exchange for the countertransport
of calcium ions (Ca2+).
Pumps run backwards
Interestingly,
some pumps, like Na+/K+ ATPase and Ca2+ ATPase pumps, can be made to run
backwards.
When the normally pumped ions are allowed to diffuse backwards through
the pumps (down their diffusion gradients), the energy molecule ATP is
reconstituted (from ADP and phosphate) within the cell. The energy for
this being extracted from the ions diffusing down their diffusion gradients.
Ion channel conductances
Across the
membrane there are channels for specific ions (mainly K+, Na+, Ca++, Cl-).
Ions move across the cell membrane through these channels (when open) under
the influence of concentration gradients (chemical potential) and the electric
field, which depends on the membrane voltage. For K+ and Cl- the force
from the inward pointing E field tends to cancel (much of) the pressure
from the concentration gradient. However, for Na+ and Ca++ the E field
and concentration gradient are aligned in the same direction, both pushing
the ions into the cell. A consequence of this (I think) is that normally
Na+ and Ca++ channels must be closed, whereaas K+ channels (and maybe Cl-
channels) can (probably) remain open.
To an electrical
engineer these channels are like a bunch of resistors in parallel each
ion channel having its own resistance (conductance). The flow of ions (which
are charged) is, of course, a current. So ohms law applies, and each ion
current = (membrane voltage)/(its channel resistance). The voltage required
to stop diffusion down an ion channel is called the Nernst potential,
and its formula (for K+) at human body temperature (37C) is below.
E = - 61 mv x log (K+ inside/K+ outside)
Since the log
function is base 10, a 10:1 concentration ratio yields 61 mv and
a 100:1 ratio 2 x 61 mv = 122 mv. For a typical potassium concentration
of K+ inside the cell x20 higher than outside its Nernst potential is E
= - 61 mv x log (20) = - 79 mv.
The polarity
of the Nernst potential depends on both the polarity of the ion and on
whether the inside concentration is higher or lower than outside.
For potassium, which is a positive ion with a higher concentration inside,
the membrane voltage is negative (inside relative to outside). For sodium,
also a positive ion but with higher concentration outside, the voltage
is positive.
It turns out
that in animal cells the membrane voltage is pretty much set by the potassium
ion (K+). The reason for this is that the conductance of the K+ ion channel
is much higher (by about a factor of 20) than the other ion channels. (In
simple terms --- the potassium channels are open and the other ion channels
are closed.) This results in a cell's resting membrane voltage being usually
just a little lower (in magnitude) than the voltage needed to stop K+ from
diffusing down its ion channel and out of the cell. Typical numbers (for
a heart cell) are membrane resting potential -90 mv, whereas its K+ Nernst
potential is -96 mv. This less than complete cancellation (6 mv) leads
to a slow, but steady, 'potassium leakage' out of the cell, which is compensated
for by pumping K+ back in using the main Na+/K+ ATPase pump.
For Na+ and
Ca++ the E field and concentration gradient are aligned and pushing
inward. The flow rate of these ions is low only because these ion channels
have a very high resistance, in other words these channels are basically
closed..
Circuit model of the membrane
To me, as
an electrical guy, what I really wanted to understand the membrane voltage
was a circuit model of the membrane. It could (it seemed to me) include
pumps (sources), ion flows (currents), chemical potentials (batteries due
to concentration gradients), ion channel conductances (resistances) all
connected to the membrane capacitance. A good circuit diagram with these
elements would really show (at least to an electrical engineer) how the
voltage across the membrane capacitance is controlled, and would be especially
helpful in understanding how active cells rapidly change their membrance
voltage.
Unfortunately
nearly everything written on this subject is written by biologists, who
generally describe all this complexity only in words, making it nearly
impossible to really understand. I looked through a textbook on the eukaryotic
cell (1,000 pages), and no circuit diagram. After much searching on the
web, I finally found that membrane circuit diagrams do exist (though to
date I have found only one). It's in a Univ of New Mexico biology exam
(with answers). Here is a link to it: http://www.unm.edu/~toolson/435_samp1_2001_key.html.
Below is a membrane ciruit diagram I have drawn. It's like the Univ of
New Mexico diagram, but more complete.
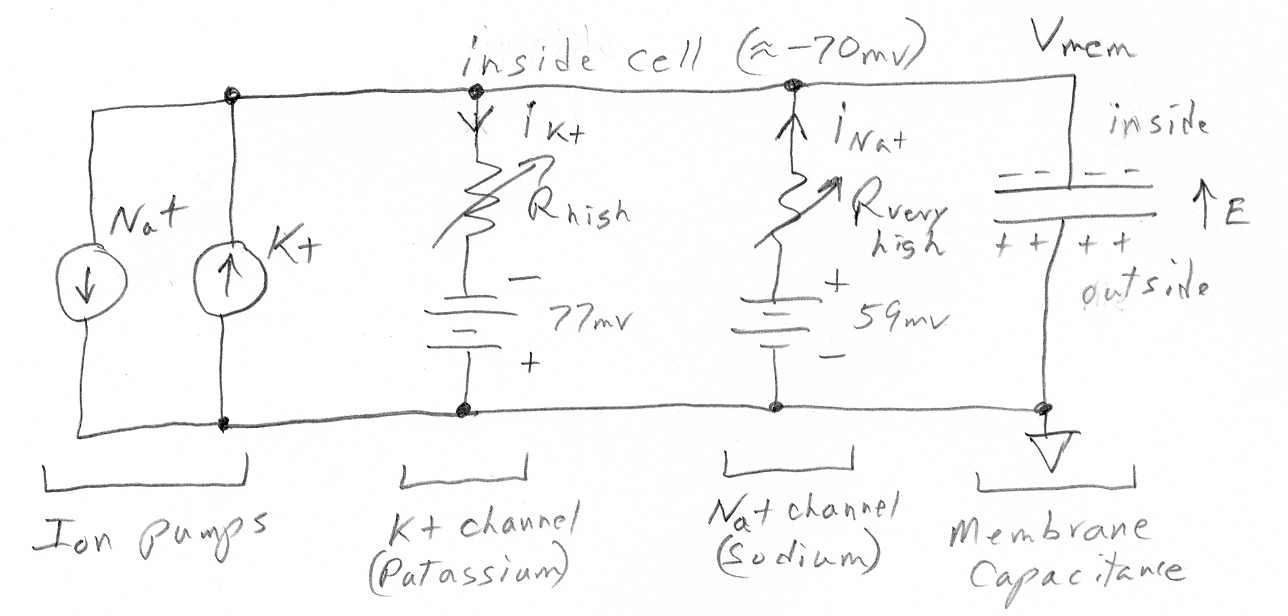
Membrane circuit diagram with K+, Na+ ion diffusion
channels and associated pumps.
The batteries in series with the resistance of the
ion channels are the Nernst potentials associated with the K+ and Na+ concentration
gradients, assumed to be 20:1 (K+) and 1:10 (Na+) (inside to outside).
More membrane circuit models
I finally
stumbled on a book reference for membrance circuit models. It's handbook
like with definitions and general equations, but nothing as detailed as
I have above. The link below opens a Biomedical Engineering Handbook (Ch11
-- Membrance Models).
http://www.cmu.edu.cn/jcyxy/upl_files/2008112218461948.pdf
My sketch above
seems to be called by the biologists a Hodgkin-Huxley resistor battery
model.
-- Hodgkin-Huxley
is the frequently used model. It models the passive flow of ions across
the membrane.
-- resistance
in the model can be non-linear and time varying (think nerve cells)
-- battery
voltage is the Nerst Potential (for the ion modeled)
E = (1/z) x (RT/F) x ln{ion concentration inside/ion concentration outside}
z is valence of ion
-- most cell
membranes are lipid and act as a capacitor (C = 1 uf/cm^2) or (1 pf for
(10um x 10um))
-- at dilute
concentrations ions in aqueous solution behave like a gas (this is why
the gas constant R appears in the equations)
Active cells
The voltage
reversal figure below is from the same Univ of New Mexico exam. This type
of cell is called an active cell because it modulates (by gating
on/off its ion channels) its cell membrane voltage. The most common active
cells are muscle and nerve cells, but interestingly the female egg cell
membrane is active too, changing the membrane potential (from negative
to positive) after a sperm enters to block further sperm. Still another
type of active cell is the photoreceptor cells of the eye where photons
trigger (via modulation of Na+ flows) cell voltage changes.
Biologists
call the voltage reversal shown in the figure (below) a membrane depolarization,which
I find a little misleading. Note it's very fast, an ion current pulse and
resulting voltage reversal can happen in less than 1 msec. The reversal
in this (unspecified) cell is caused by the cell opening positive Na+ ion
channels (blue) in its membrane. This allows a large pulse of N+ ions to
flow into the cell (down the Na+ concentration gradient). This (positive
ion) current charges the membrane capacitance causes the cell membrane
voltage to change from -65 mv to +40 mv. A msec or so later as the sodium
channels are closing, the cell opens potassium ion (K+) channels (green)
for a few msec. This allows potassium ion (K+) current to flow out of
the cell (down the K+ concentration gradient) restoring the original negative
charge on the membrane capacitance. (In some cells like heart muscle cells
there is a delay of a msec or two before the K+ ion channel opens to repolarize
the cell making the voltage pulse wider than shown in this figure.).
These ion flows
reduce the K+ and Na+ concentration gradients, but only a little, and over
time they are restored to their original concentrations by the K+/Na+ pumps
in the membrane (pumping Na+ out and K+ in). (Wikipedia at this link (http://en.wikipedia.org/wiki/Membrane_potential)
works some back of the envelope numbers showing that the change in concentration
gradients is only about 0.1%.)
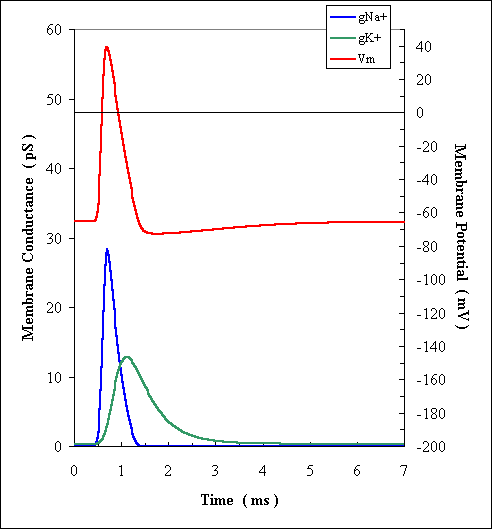
Membrane transient voltage reversal (depolarization)
Blue & green pulses in the figure are not (ion)
currents, but changing conductance (inverse of resistance) of the ion channels.
The ion current pulses (not shown) are, however, basically similar, except
the pulse shapes are somewhat different (due to the changing voltage).
(Univ of New Mexico biology exam, 2001, Toolson)
From the curves
above we can work some numbers. The charge flows of K+ and Na+ must balance
since the membrane voltage starts and ends at the same voltage. The current
is equal to the conductance x voltage (version of ohms law). Looking at
the blue curve above we can make estimates of current, charge, and capacitance.
Current is the (rectangular equivalent) voltage (across
the Na+ conductance) x (rectangular equivalent) conductance.
i = (av) voltage x (av) conductance
= 1/2 x (65 + 45) mv x 1/2 x 28 x 10^-12 x siemen
= 0.77 x 10^-12 A
(0.77 pa)
q = current x time
= 0.77 x 10^-12 amp x 0.8 msec
= 0.62 x 10^-15 coulomb
Membrance capacitance is charge flow divided by voltage
change
C = charge/voltage
= 0.62 x 10^-15 coulomb/(65 +40) mv
= 5.9 x 10^-15 farad (5.9 ff or femto farad)
One coulomb is the amount of electrical charge carried
by 6.24 x 10^18 electrons (or single ions), so the number of ions flowing
into (and out) of the cell during each pulse
# of ions = q x 6.24 x 10^18 ions/coulomb
= 0.62 x 10^-15 coulomb x 6.24 x 10^18 ions/coulomb
= 3.87 x 10^3 ions
= 3,870 ions
Check --- Maybe. Kimball's online biology reference says
a measured value for (some type of) nerve cell is 7,000 Na+ ions in one
msec for one
ion channel. So it could be that the vertical scaling
in the figure above is for a much smaller cell than Kimball referenced
(very possible, since large cells are usually measured), or it might be
for only one channel, or it might just be wrong.
In rectangular equivalent time (0.8 msec) the Na+ charge
flows in, so the Na+ ion (diffusion) flow rate is
Na+ ion flow rate = # of ions/0.8 msec
= 3,870 ions /0.8 msec
= 4.8 million ion/sec
How cells communicate
For nerve
signals to propagate and muscles to contract adjoining nerve cells and
muscle cells need to 'talk' to one another. They (apparently) do this by
flipping (pulsing) their membrane voltages. Sections of the membrane in
the same cell communicate this way too.
The (Na+) ion
flow channels across the membrane respond directly to the voltage of adjoining
regions. When a section of membrane 'sees' a voltage more positive than
its threshold voltage (about -50 mv in mammal cells), it opens briefly
its Na+ ion channels allowing diffusion down concentration gradients, driving
the cell voltage fully positive (+40 mv). The biologist speak of this as
a "wave of (de)polarization" sweeping across the membrane.
Voltage 'flip'
Cell voltages
in active cells are able to change rapidly (1 msec or so) by allowing transient
ion flows across concentration gradients previously set up in the cell
by ion pumps. Then there is a recovery period (refractory period) while
the pumps restore the original ion concentrations. The voltage 'flip' is
really a three step process. First an ion flow drives the cell voltage
up,
then immediately following a second (different) ion flow drives the voltage
back down again. Thirdly, there is a recovery time while all the (slightly)
disturbed ion concentrations are restored by the cell pumps.
To 'flip' the
voltage up (drive it positive) one (or more) of the positive ions that
have been pumped out of the cell are allowed to flow back in. The candidates
are sodium (Na+) and calcium (Ca2+). To do the second part of the 'flip',
i.e. drive the membrane voltage negative again, some of the positive potassium
ions (K+), whose concentration is pumped up in the cell, can be allowed
to diffuse out, or the negative ion chloride (Cl-), whose concentration
is low inside the cell, can be allowed to diffuse in. In practice in nearly
all active cells it is flows of Na+ in followed by K+ out
that cause the voltage flip, which is just what is shown in the figure
above. This is nicely consistent with the fact that the major pump in cell
membranes is a combo Na+ (out) and K+ (in) pump, called the Na+/K+ ATPase
transporter.
To put some
numbers on it (see above) a voltage 'flip' is a fast ion flow (diffusion)
into the cell of (about) 1 pa for 1 msec of positive Na+ ions, which changes
the cell membrane voltage (about) +100 mv (from -70 mv to +40 mv), followed
by a similar ion pulse flow out of the cell of positive K+ ions. Recovery
time (refractory period), where the Na+/K+ pump restores the concentration
gradients, is typically another msec or two. Here is a figure of an active
cell voltage 'flip' from Wikipedia.
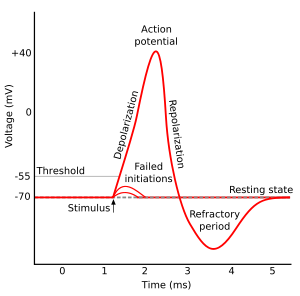
In muscle cells
the time it takes for the cell to 'twitch' (contract & relax) is 50
msec or so, much longer than the time it takes for its membrane voltage
'flip' and its ion concentrations to recover (refractory period), hence
rapidly stimulating the cell (every 20 msec or so) can keep a muscle cell
fully contracted. A sustained muscle contraction caused by rapid triggering
goes by the weird name of 'tetanus'. It's the opposite for heart
cells, refractory (recovery) time exceeds twitch/contraction time, so they
cannot stay contracted. (One reference comments that this is a good thing!)
Nerve cells
Nerve cells
normally have a negative voltage across their membranes too (-70 mv or),
but unlike other cells they are able to change their membrane voltage rapidly
and to do so when the cell is stimulated in some way externally. This is
the mechanism they use to transmit information (on/off, binary information)
from one nerve cell to another nerve cell.
Nerve cells
have many ion channels that are gated, i.e. they can be triggered open
or closed. When these ion channels trigger open (in sequence), the voltage
rises and then falls quickly ('flips') in a msec or so. This is not because
the ion concentrations change much, but because the conductivity (resistance)
across the membrane is modulated. When Na+ channels open strongly, the
membrane voltage is driven toward the Nernst potential of Na+. This is
positive (reversed from K+), because the Na+ concentration in the cell
is low. A cell with a collapsed (or reversed) membrane voltage is said
to be 'depolarized'. This depolarization can be sensed by adjoining nerve
cells, allowing signals to propagate from nerve cell to nerve cell through
the body.
Light sensitive cells
Like all cells
the photoreceptor cells of the eye have pumps which pump up the
concentration of K+ (inside the cell) and pump down the concentration
of all other ions like Na+ and Ca2+. In the dark photoreceptor cell membranes
have open channels for K+ (ungated) and Na+ (light gated off) and Ca2+
(voltage gated). Since these channels are to some degree (in the dark)
open, the ions diffusing (and drifting) down their channels are recycled
and concentrations maintained by a high density of K+/Na pumps.
Wikipedia calls
photoreceptor cells "strange" because in their dark state (normal state)
the cell voltage is 'depolarized' (due to the leaky Na+ ion channels) causing
the cell voltage to be (partially) collapsed at -40 mv. Photon hits trigger
the Na+ and Ca2+ channels to close allowing the cell voltage to
fully polarize (or hyperperpolarize, which just means to increase the potential)
to -70mv, set by the open, ungated, K+ channels. It is this light induced
voltage change that activates the next cell and sends an excitatory signal
down the neural pathway. The Na+ ion diffusion flow into a dark cell is
called the 'dark current'. In an (unspecified) time after a photon hit,
the Na+ channels reopen, the cell voltage drops back to its depolarized
-40mv, and the cell is ready for another photon hit.
Ion transport controls a cell's water pressure
Control of
ion concentration inside cells indirectly (via osmosis) controls water
pressure in the cell. Osmosis is a special term used for the diffusion
of water through cell membranes. Water is never transported actively; that
is, it never moves against its concentration gradient. Water passes by
diffusion from a region of higher to a region of lower concentration, meaning
the concentration of water.
When ion pumps
and diffusion channels produce a high concentration of ions inside the
cell (relative to outside), it means the concentration of water inside
the cell is lowered, so water will diffuse into the cell (building
up pressure). Conversely low ion concentration inside causes water in the
cell to diffuse out of the cell, lowering pressure.
Ion channels compared to pn junctions
Inside the
common silicon diode there is a zone (pn junction) not unlike the ion channels
in cell membranes. Highly doped silicon is electrically neutral, but has
an interesting property. One polarity of charge is mobile and the other
polarity is immobile, and (importantly) in p and n type silicon which charge
type is mobile and which fixed is reversed. So at the interface between
the p and n materials (pn junction), a diffusion gradient exists tending
to cause net diffusion of mobile charges across the pn junction. When some
of the mobile charges do diffuse over to the other side, however, the fixed
charges in the silicon are (in effect) exposed and an electric field develops
across the pn junction that opposes further diffusion.
The diffusion
gradient in silicon is far higher than in cells, so the potential across
pn junction is typically 800 mv (about x10 higher than across cell membranes),
however, the basic mechanism generating the voltage (mobile and
fixed charges self-cancelling a diffusion gradient) is the same as
it is in the cell.
Footnote ---- A diode just sitting on a
table, totally unpowered, has internally an electric field with 800 mv
potential across its pn junction. (An unpowered diode is like a 'resting'
cell.) The flow of charge talked about above essentially occurs once when
the diode is manufactured. When the diode is used in a circuit, slight
modulations externally of the voltage across the junction control current
flowing (by diffusion) through the diode.
The analogy
is made that the voltage across a pn junction (or cell membrane) is like
a height of dam with a spillway. Slight modulations in the height of the
dam (voltage) cause large variations in the flow of water (charge) through
the spillway.
Cell membranes (Wikipedia excerpts)
The cell membrane
is a semipermeable lipid bilayer common to all living cells. The movement
of substances across the membrane can be either passive, occurring without
the input of cellular energy, or active, requiring the cell to expend energy
moving it across the membrane. For determination of membrane potentials,
the two most important types of membrane ion transport proteins are ion
channels and ion pumps.
Ion channel
proteins create paths across cell membranes through which ions can passively
diffuse without expenditure of energy. Most cells have potassium-selective
ion channel proteins that remain open all the time.
While most
descriptions of the genesis of membrane potential begin with the concentration
gradients already in place, as if by magic (therefore ignoring the principle
of conservation of energy), these gradients can only be created at the
expense of putting energy into the system. This work is done by the ion
pumps (ion transporters or exchangers) and generally is powered by
ATP. The resting membrane potential is not an equilibrium potential
as it relies on the constant expenditure of energy (for ionic pumps as
mentioned above) for its maintenance.???
One of the
key roles of the membrane is to maintain the cell potential. In most cells
the resting potential has a negative value, which by convention means that
there is excess negative charge inside compared to outside. For most animal
cells potassium ions (K+) are the most important for setting the resting
potential. Due to the active transport of potassium ions, the concentration
of potassium is higher inside cells than outside. (Note, the high concentration
of positive potassium ions inside the cell is (initially) charge balanced
by immobile?? negative (An-) ions (free electrons?)
The outward
movement of positively-charged potassium ions is due to random molecular
motion (diffusion) and continues until enough excess positive charge accumulates
outside the cell to form a membrane potential which can balance the difference
in concentration of potassium between inside and outside the cell. 'Balance'
means that the electrical force (qE) that results from the build-up of
ionic charge increases until it is equal in magnitude but opposite in direction
to the tendency for outward diffusive movement of potassium.
The typical
membrane potential of a cell arises from the separation of potassium ions
from intracellular immobile (meaning unable to move through the ion channels?)
anions across the membrane of the cell. If the membrane were to become
permeable to a type of ion that is more concentrated on one side of the
membrane, then that ion would contribute to membrane voltage because the
permeant ions would move across the membrane with net movement of that
ion type down the concentration gradient.
(Wikipedia
figure shows) K+ diffusing out of the cell. (initially inside of
cell is neutral with as many K+ and An- ions) As positive charge builds
up outside the membrane and negative (due to loss of K+) inside the membrane
an E field pointed inward develops. This E field pushes K+ ions back toward
the inside of the cell. Steady state is reached when the E field is strong
enough to bring (net) K+ diffusion across membrane to a halt. (energy
is transferred from the concentration gradient to the electrical energy
(voltage) across the membrane capacitance).
What's the purpose of the cell potential?
While cells
expend energy to transport ions and establish a transmembrane potential,
they use this potential in turn to transport other ions and metabolites
such as sugar. (The cell E field will pull any positive ion
from the outside into the cell if the cell provide an open channel for
the ion.)
------------------------------------
Electrochemical gradient
-- electrochemical
potential is the mechanical work done in bringing 1 mole of an ion from
a standard state to a specified concentration and electrical potential.
-- In
mitochondria and chloroplasts, proton gradients are used to generate
a chemiosmotic potential that is also known as a proton motive force. This
potential energy is used for the synthesis of ATP by oxidative phosphorylation
- An electrochemical
gradient has two components. First, the electrical component is caused
by a charge difference across the lipid membrane. Second, a chemical component
is caused by a differential concentration of ions across the membrane.
The combination of these two factors determines the thermodynamically favorable
direction for an ions movement across a membrane.
-- With respect
to a cell, organelle, or other subcellular compartments, the inclined tendency
of an electrically charged solute, such as a potassium ion, to move across
the membrane is decided by the difference in its electrochemical potential
on either side of the membrane, which arises from three factors:
* difference in the concentration of the solute between the two sides of
the membrane
* charge or "valence" of the solute molecule
* difference in voltage between the two sides of the membrane (i.e. the
transmembrane potential)
-- A solute's
electrochemical potential difference is zero at its "reversal potential".
The transmembrane voltage to which the solute's net flow across the membrane
is also zero. This potential is predicted theoretically either by the Nernst
equation (for systems of one permeant ion species) In physiology
the Nernst equation is used for finding the electric potential of a cell
membrane with respect to one type of ion.
-- The potential
level across the cell membrane that exactly opposes net diffusion of a
particular ion through the membrane is called the Nernst potential for
that ion.
-- Nernst equation
for cell membrane voltage. Note 'ln' arises because the ratio of oxidized
to reduced molecules, [Ox]/[Red], is equivalent to the probability of being
oxidized (giving electrons) over the probability of being reduced (taking
electrons). And these are describe with Bozmann statistics e^-(volt
treshold/KT) . Also -- Dividing by e converts from chemical potentials
to electrode potentials, and (kT/e) = (RT/F)
E = E0 - (RT/nF) ln (reduced concetration/oxided concentration)
+ (59mv/n) log (ion outside of cell/ion inside of cell)
Osmosis overview
Inside a cell there is water with stuff (larger molecules) dissolved in
it. The small water molecule is able to diffuse though the cell wall and
membrane. If the cell is placed in an water with less stuff dissolved in
it (it doesn't matter what the stuff is!) and the pressure inside the cell
is the same as outside, what happens is that net water molecules travel
into
the
cell raising the pressure of the cell against its cell wall.
Water diffuses
into
the cell, because inside the cell fewer water molecules per sec hit the
barrier than on the outside since inside the water molecules are spaced
further apart due to the stuff dissolved in it. (From an entropy point
of view the water moves to try an equalize the solute concentrations.)
As water diffuses in, the pressure inside the cell builds up, eventually
stopping the net inflow of water. (The higher pressure must either
increases the speed of the water molecules or the number of water molecules/sec
hitting the barrier.)
Osmotic pressure
a very important to cells. For example, it builds up high pressure in the
cells of plant leaves giving the leaf structure. If you drink sea water,
(apparently) there is more salt in the sea water than solutes in your cells,
so water diffuses out of your cells. Reverse osmosis, which is used to
desalinate sea water, is simply applying pressure higher than the osmotic
pressure to cause water to diffuse from the side with lots of solutes
to the side with fewer.
-- (osmosis)
Water is never transported actively; that is, it never moves against its
concentration gradient. However, the concentration of water can be altered
by the active transport of solutes and in this way the movement of water
in and out of the cell can be controlled.
Energy conversion
Reaction energy
is often given in Kcal/mole (where mole = 6 x 10^23) or Kj/mole. These
can be converted into electron-volts. (1 Kcal = 4.2 x 10^3 joule) (1 Kj
= 1 Kcal/4.2) (A mole of photons (6.2 x 10^23) is called an Einstein.)
100 Kcal/mole = 100 x 4.2 x 10^3 joule}/(6.2 x 10^23) = 68 x 10^-20 joule
1 ev = 1.6 x 10^-19 joule
68 x 10^-20 joule /1.6 x 10^-19 = 4.25 ev
so
100 Kcal/mole <=> 4.25 ev
100 Kj/mole <=> 1 ev
-- Cells need
energy to drive reactions. The molecule that supplies the energy is ATP
(This reaction is called ATP hydrolysis). When the third phosphate is removed
by hydrolytic cleavage, 7 kcal of energy is released per mole of ATP.
ATP + H2O --->
ADP + Phosphate + Energy (7 kcal)
When the second phosphate is removed, the same amount
of energy is released.
ADP + H2O ---> AMP + Phosphate
+ Energy (7 Kcal)
The bonds between
the two phosphates are not strong bonds. In fact, these bonds are
easily broken releasing 7Kcal of energy per mole. 7 Kcal of energy
is enough to drive endergonic reactions in the cell. All the energy does
not come from the moving of electrons to a lower energy level. In fact,
the rearrangement of electrons in other orbitals (i.e..ATP ---> ADP) result
in a structure with less energy.
ATP
ATP is made
up of the nitrogenous base adenine, the five-carbon sugar ribose and three
phosphate groups. Three phosphate units (triphosphate), each made up of
one phosphorus atom and four oxygen atoms, are attached to the ribose.
The two (covalent) bonds between the three phosphate groups are relatively
weak and yield their energy readily when split by enzymes.
Wikipedia formula
--- C10 H16 N5 O13 P3
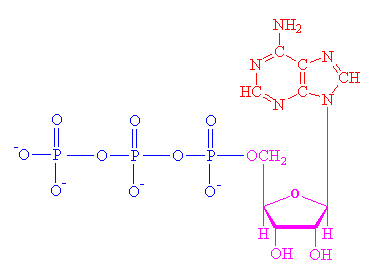
ATP consists of three phosphates (blue), a
sugar, ribose (magenta), and a nitrogen base, adenine (red).
Removal of the left phosphate (PO4) converts ATP to
ADP and releases energy.
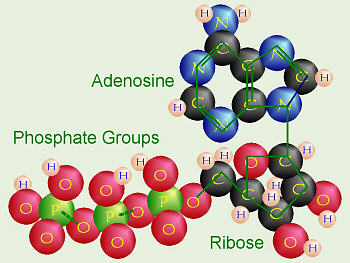
ATP, well it almost agrees with stick figure above
Count here agrees with Wikipedia formula --- C10 H16
N5 O13 P3
Notice ribuse here has 8 H whereas in figure above
there is 6 H
source -- http://biology.clc.uc.edu/Courses/bio104/photosyn.htm
Inside a cell
the ATP molecule is split at one of the high energy bonds, releasing the
energy to power cellular activities. Adenosine diphosphate (ADP) and phosphorus
(P) are produced in the process. With the release of the end phosphate
group, 7 kilocalories per mole (under laboratory conditions, about 11 kilocalaries
per mole in cell) of energy become available for work.
ATP + H2O --> ADP + Phosphate + energy
ATP needs to
be regenerated continuously by the recombining of ADP and P. But note the
synthesis of ATP requires that ADP be available. The burning of food to
make ATP is regulated by the levels of 'spent' ATP (ADP) in the cell. ATP
is made only as needed, the process is self-limiting.
----------------------------------------------------------------------------------
(atomic bonding isn't really cell biology. Does this
below with Atoms, or its own essay?)
Atomic Bonding Overview (4/08)
All chemical
energy (including explosives) comes from a rearrangement of (shared) electron
bonds that changes their potential energy. Potential energy lost is released,
and conversely potential energy gained must be input to drive the reaction.
Pairs
Electrons
always want to pair up (one spin up, one spin down), it's the heart of
bonding. An up spin electron can share the same space as a down
spin electron. When electrons pair up by forming covalent (etc) bonds,
the electrons lose potential energy and this energy is released.
Pulling (shared) electrons toward the nucleus -- Electronegativity
Electrons
that move closer to a nucleus lose potential energy, so energy is released.
This is the 'secret' of how reactions with atoms that have high electronegativity,
like oxygen, put out energy. In other words why burning things makes heat!
Electronegativity
is measure of how strongly a (shared) electron pair is attracted to an
atom's nucleus. The cause of electronegativity is really quite simple.
It's a measure of the electrical attraction between the fraction of the
positive nucleus that is unshielded and the shared (negative) electron
pair. It's higher if the nucleus has more protons and higher if the (shared)
electrons are in a lower orbit, and thus closer to the nucleus.
And -- key
concept -- if (shared) electrons do in fact end up closer to the
nucleus, then (net) energy is released as the potential energy of the electrons
drops.
* Fluorine (element 9) has the highest electronegativity of any element
in the periodic chart. The shared electron pair are in level 2 and they
'see' a (net) positive charge of 7 ( = 9 protons - 1s inner pair shielding).
Electronegative value = 3.98.
* Oxygen (element 8) electronegativity is high, but not quite as high as
fluorine because it's (net) positive charge is 6 ( = 8 protons - 1s inner
pair shielding). Electronegative value = 3.44.
* Chlorine (element 17) shared pair, like fluorine, also see a net positive
charge of 7 (= 17 protons - 10 electron shielding in levels 1 and 2), but
the shared pair is in level 3 so it's a little further from the nucleus
than in fluorine, this the attraction is a little weaker. Electronegative
value = 3.16
* Hydrogen (element 1) has a very low net positive charge (1) because it
only has one proton, but it's electronegativity is increased by the fact
that the shared electrons 'orbit' quite close to the nucleus in the 1s
orbit. Electronegative value = 2.2.
How oxygen bonding (oxidation) releases (net) energy
When oxygen
is able to pull the shared electrons it wants from another atom with a
lower electronegativity (which includes carbon and hydrogen and just about
every other atom), it pulls the shared electrons in closer to its nucleus.
This lowers their potential energy, and this energy is released. Of course,
to get an unattached oxygen atom (from the atmosphere) some energy must
be expended, since oxygen in air is already bonded to another oxygen (O2),
so an oxygen-oxygen covalent bond (of molecular oxygen) needs to be broken.
But the energy needed to break an oxygen-oxygen covalent bond is not as
high as will later be released when the oxygen combines with carbon or
hydrogen. The reason is an oxygen bond in O2, being between identical atoms,
is symmetrical; it's a true covalent bond. The shared electrons are located
(on average) right between the two oxygen nuclei. Here's the electronegativity
values of oxygen, carbon, and hydrogen.
Oxygen
3.44
Carbon
2.55
Hydrogen
2.20
There's actually
a pretty linear relationship between electronegativity and bond energy.
The graph below shows that making bonds with oxygen typically outputs
about 40% more energy than is required to break carbon and
hydrogen bonds and the oxygen-oxygen bond too (2,000 vs 1,300 -1,500 KJ/mole).
Vertical scale is the usual Pauling electronegativity values (from 0.7
to 4) and horizontal is bond energy in KJ/mole.
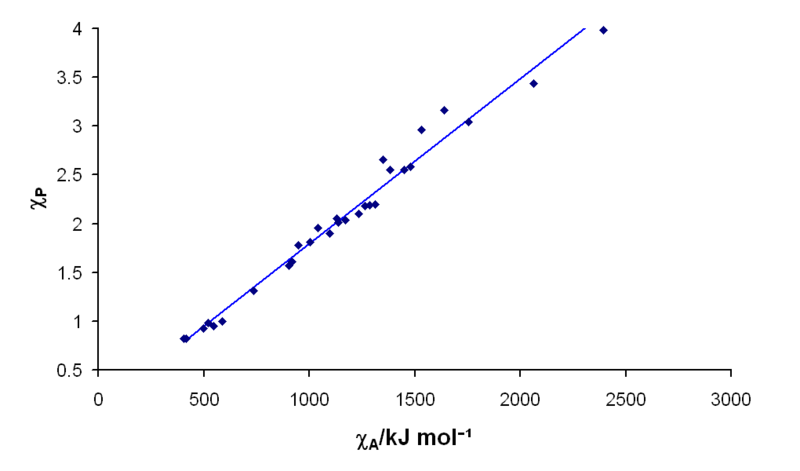
(from Wikipedia -- Electronegativity)
Working some numbers --- Oxidation of methane
Methane (CH4)
is the simplest hydrocarbon. It's just one carbon surrounded by four hydrogen,
each hydrogen attached to the carbon by a single covalent bond. Since the
electronegativity of carbon is a little higher than hydrogen (2.66 vs 2.2),
the shared electrons are a little closer to the carbon nucleus than the
hydrogen.
Burning methane
(in air) yields CO2 and water. The reaction breaks the oxygen-oxygen covalent
bonds, then half of the oxygen bonds covalently with the hydrogen (making
water, H2O) and the other half with the carbon (making carbon dioxide,
CO2). Here's the chemical formula (you adjust quantities so atoms out =
atoms in):
CH4 + 2 O2 = CO2 + 2 H2O
To find the
(net) energy released in the burning of methane (to what degree of accuracy?)
you just find the energy released in the newly made bonds (on the right)
and subtract off the energy required to break the bonds (on the left side).
Atomic bond energies are tabulated. (a double covalent bond is two shared
electrons whereas a single covalent bond is one shared electron)
CO2
2 O=C double covalent
2 x 799
2 H2O
4 O-H single covalent
4 x 460
CH4
4 C-H single covalent
- 4 x 410
2 O2
2 O=O double covalent
- 2 x 494
------------------------------------
3438 - 2628 = 810 kJ/mole (released)
So adding (&
subtracting) the bond energies tells us that burning a mole of methane
(16 grams) releasees 810 kJ = (194 kcal/mole, 8.39 ev) of energy.
Some useful molecule total bond energies:
H2O
2 O-H single covalent 2
x 460 = 920 Kj/mole = 220 kcal/mole = 9.53 ev
CO2
2 O=C double covalent 2 x 799 = 1,598
Kj/mole = 382.5 kcal/mole = 16.56 ev
O2
1 O=O double covalent 1 x 494 = 494 Kj/mole
= 118.3 kcal/mole = 5.12 ev
H2
1 H-H single covalent 1
x 460 = 460 Kj/mole = 110 kcal/mole = 4.76 ev
Covalent, polar, & ionic
A covalent
bond is just a shared electron pair. If the electronegativity of the two
atoms are (significantly) different, i.e. if the electrostatic 'pull' of
the two nuclei are (significantly) different, then the electron pair moves
closer (on average) to the stronger pulling nucleus.
* The electron pair moving closer to one nucleus makes that atom acquire
a (net) negative charge and the other atom (from which the electron pair
is more distant) acquires a (net) positive charge. A molecule with positive
and negative atoms is called a polar molecule, and these plus and
minus 'ends' causes some attraction between molecules. Water is polar and
each water molecules pulls on four others. This raises its boiling point
because (extra) heat is required to break all molecule to molecule (polar)
attraction.
* If the electron pair is much closer to one nucleus than another, then
the covalent bond is called ionic. The classic case is NaCl. The
shared electron pair is in level 3. Sodium (element 11) on the left side
of the periodic chart only has a (net) positive attraction of 1 ( = 11
protons - 10 electron shielding in levels 1 and 2), whereas chlorine on
the right side of the periodic chart has a (net) positive charge of 7 (=
17 protons - 10 electron shielding in levels 1 and 2). When salt disolves
in water, the sodium and chlorine separate with chlorine keeping the extra
electron resulting in ions Na+ and Cl-.
* (I think) Pulling of the shared electron pair closer to one nucleus (quasi-ionic)
also reduces the electron's potential energy and thus releases energy.
This link is
by far the best bonding tutorial I have found. Very clear discussion of
orbit changes of many different bond types (worth studying in detail):
http://www.chemguide.co.uk/atoms/bondingmenu.html#top
----------------------------------------------------------------------------------
(This is an earlier treatment of the same material when
I understood it less well)
Atomic bonds -- Electronegativity
Atoms bond
into molecules by overlapping their electronic orbitals. Whether this is
called 'sharing' or 'donating/accepting' electrons, it's really the
same thing. Wikipedia link on Electronegativity (see below) is a good introduction
to atomic bonding. Electronegativity is a measure of the ability
of an atom or molecule to attract electrons in the context of a
chemical bond. The link has a periodic chart showing the electronegativity
value of every element (ranges from about 1 to 4) with oxygen at 3.44 having
the 2nd highest value (fluorine is highest at 3.98). (Electronegativity
is so important even my supermarket sells it! Yes indeed, I was able
to buy a periodic chart with all the negativity values in my supermarket
-- part of a plastic 'crib notes' series for students).
When bond is
made between only two atoms, it's typically an atom from the left side
of the periodic table, which have a few excess (outside a full shell) electrons,
joining an atom from the right side of the table, which are just a few
electrons shy of completing an electron shell. When multiple atoms bond,
there are a lot of different ways to fill a shell. For example, in CH4
(methane) carbon gets the four extra electrons it needs to fill its shell
by sharing with four hydrogen. From the hydrogen perspective it gets the
one extra electron it needs to fill its shell by sharing one of the carbon
electrons. (Note, shell filling is just a set of 'rules of thumb'
that make it easy to remember the lower energy orbit states.)
Atomic bonds -- Covalent vs ionic
There are two (idealized)
types of chemical bonds, Covalent and Ionic, but really they
shade into each other with the key parameter being the
relative
electronegativity. This is nicely shown pictorially in this link:
http://www.chem.ox.ac.uk/vrchemistry/electronsandbonds/bondsperatom1.htm
Covalent ---
If electronegativity values are close, in a sense the atoms pull
on (shared) electrons equally. When the difference in the relative electronegativity
is not too great (< 1.7), the electron bond formed is called a covalent
bond. This type of bond is conventionally described as the atoms sharing
electrons. If the relative electronegativity values is < 0.4, then the
bond is balanced, so these are non-polar covalent bonds. If the
relative electronegativity values is 0.4 to 1.7, then the bond is unbalanced,
so these are polar covalent bonds. Obviously bonds between identical
atoms, like diatomic oxygen, are perfectly balanced, so these bonds are
pure (non-polar) covalent.
Ionic --- If
electronegativity values are far apart ( > 1.7), then one atoms pulls
on (shared) electrons much harder than the other. This type of bond is
called an ionic bond. This type of bond is conventionally described
as one atom donating an electron to another atom that accepts
it. The force between the atoms created by this bond is then spoken of
as being an electrostatic attractive force between the positive (donating)
ion and the negative (accepting) ion.
A dipole moment
is an electrical force that is generated because of the unequal distribution
of the bonding electrons between the two bonded atoms. In the case of an
ionic bond that unequal distribution is extreme. The dipole moment of an
ionic bond is quite large compared to polar bonds of the covalent bond.
Ionic bonds (can be) about as strong as covalent bonds.
Salt (sodium
chloride) is a classic ionic bond. Most metals (like sodium) have only
a few valence electrons hence relatively low ability to attract electrons
and low electronegativity values. Sodium (atomic number 11) has only one
valence electron, so it has a very low electronegativity value of 0.93.
Many non-metals like nitrogen, chlorine, oxygen, fluorine have nearly full
shells missing one or two electrons (nitrogen is missing three), so have
high electronegativity values. Chlorine (atomic number 17) has a seven
valence electrons, so it has a very high electronegativity value of 3.16.
When salt dissolves
in water, the sodium and chlorine atoms come apart (why? see below)
as charged ions, so here (at least) the sharing & loss of the electron
must be 100%.
--
Hydrogen and oxygen in water are covalently bonded. Water is a polar molecule,
because it has partial positive and partial negative ends. The hydrogen
atoms of the water molecule can now form bonds with other slightly negative
(polar) compounds. Each hydrogen of this water molecule can form hydrogen
bonds with oxygen atom of other water molecules. Hydrogen bonds are 20
times weaker than covalent bonds. But hydrogen bonding between molecules
is very important with organic compounds.
-- Water is
able to dissolve anything polar due to polarity. Water separates ionic
substances. Many covalently bonded compounds have polar regions, the covalent
compounds dissolve in water and are called hydrophilic (water loving) compounds.
Nonpolar substances do not dissolve in water and are called hydrophobic
(water fearing).
-- Due
to hydrogen bonds water is attracted to itself (more than the air above
it) forming surface tension. To change water from a liquid to a gas (boil
it), a lot of energy must be added to break the hydrogen bonds between
the water molecules.
-- The bond
strengths are expressed in terms of energy (kilocalories or kilojoules
per mole) that must be supplied to break the bonds under standard conditions
of temperature and pressure.
-- Carbon
backbone Carbon can form covalent bonds directly with one to
four atoms since it as four valence electrons. In many biological molecules
carbon atoms form long chains. Carbon is unique in that it can form single,
double, and triple covalent bonds with itself and other atoms.
http://en.wikipedia.org/wiki/Electronegativity
Nitrogen
Nitrogen has
two stable isotopes: N14 (7 neutrons, 99.6%) and N15 (8 neutrons, 0.4%).
Nitrogen is atomic number 7, between carbon (6) and oxygen (8). It has
five electrons in its outer shell and is therefore trivalent in most compounds.
Nitrogen is a nonmetal, with an electronegativity of 3.0.
The triple
bond in molecular nitrogen (N2) is the strongest in nature.
The resulting difficulty of converting (N2) into other compounds, and the
ease (and associated high energy release) of converting nitrogen compounds
into elemental N2, have dominated the role of nitrogen in both nature and
human economic activities.
Oxidation
Oxidation
is a slippery term. From its name you would think it means adding oxygen,
but this is only one type of oxidation. Oxidation is also used in a loose
sense to mean 'burn'. But there is an electrochemistry definition and it
appears to be totally different. So what is oxidation really? And why does
oxidizing something yield energy?
(My thinking
now is this)
Oxidize
--- A molecule is oxidized when an atom from the right side of the periodic
table (an oxidant like oxygen, fluorine, or chlorine) bonds to the molecule
causing electrons to be sucked (to some degree) from the molecule
to the oxidant atom using them to fill the oxidant's shell. The energy
stored in the combined molecule is less than the sum of its parts (due
to the filled shell), so this reaction (always?) puts out energy.
Oxidation example:
In a fuel cell gaseous oxygen (O2) is used to oxidize gaseous hydrogen
(H2) forming water. This puts out useful work in the form of electricity
+ some heat.
Reduce
--- A molecule is reduced when an atom from the left side of the periodic
table (a reductant like hydrogen, or sodium) bonds to the molecule causing
electrons to be added (to some degree) to the molecule from the
reducer. The energy stored in the combined molecule is more than the sum
of its parts (even though the reductant is down to a filled shell), so
this energy need to added to make this reaction go.
Reduction example:
Electrolysis of water into gaseous oxygen and hydrogen. This is the reduction
of hydrogen (putting back the electrons that the oxygen sucked away), and
for it to occur energy must be added in the form of running electricity
through the water.
As stated in Wikipedia:
"Oxidation
is the loss of electrons by a molecule, atom or ion"
"Reduction
is the gain of electrons by a molecule, atom or ion"
When a carbon
atom 'loses' (shares) two electrons to each of two oxygen atoms it bonds
with, Wikipedia speaks of this as 'oxidation' of carbon to yield CO2. Yet
when carbon joins with four hydrogen, Wikipedia refers to this as 'reduction'
of carbon by hydrogen to yield methane (CH4).
Is it oxidation of
carbon when it joins with an atom with a higher electronegativity value
(like oxygen), and reduction when it joins with an atom with a lower electronegativity
value (like hydrogen)?
Yes!
Here (I think) is the key --- When electrons are shared, they are pulled
spatially toward the atom with the higher electronegativity value. So higher
electronegativity atoms (oxidants) suck away electrons from the bonding
molecule, and it is the loss of electrons by the molecule that is (technically)
the definition of oxidation. It is correct (if confusing) to speak of carbon
being reduced by hydrogen, because carbon being the higher electronegativity
atoms sucks the electrons from the hydrogen to itself, thus technically
since carbon has gained electrons it has been reduced! It's clearer I think
to speak of it as carbon oxidizing the hydrogen.
element
electronegativity
-------------
---------------------
oxygen
3.44
carbon
2.55
hydrogen
2.20
My guess is
that oxygen's so-called high affinity for attracting electrons (its high
electronegativity value) means that the oxygen atom when it bonds with
other atoms that bring two electrons to share with it, drops into a lower
energy state and (somehow) releases energy in the form of heat and useful
work like current (fuel cells), electrons flows, and ion pumping.
My model
is gravity. When an object is attracted to a body by gravity, energy is
released either as useful work or as heat. Is the oxygen atom like this?
Is energy released when two electrons 'fall into' orbitals of oxygen allowing
it to complete its electron shell?
Wikipedia excerpts on oxidation/reduction
-- Oxidant
removes electrons from another substance, and is thus reduced itself.
(mnemonic) "OIL RIG" -- Oxidation Is Loss, Reduction Is Gain.
-- Oxygen is
an oxidant, but not the only one. Despite the name, an oxidation reaction
does not necessarily need to involve oxygen.
-- The gain
of oxygen, loss of hydrogen and increase in oxidation number is also considered
to be oxidation, while the inverse is true for reduction.
-- Oxidants
are usually chemical substances with elements in high oxidation numbers
(e.g., H2O2, MnO4-, CrO3, Cr2O72-, OsO4) or highly electronegative substances
that can gain one or two extra electrons by oxidizing a substance (O, F,
Cl, Br).
-- The oxidation
state of an ion is the number of electrons it has donated (+) or
accepted (-) compared to its neutral state (which is defined as having
an oxidation state of 0).
-- A reaction in
which both oxidation and reduction is occurring is called a redox reaction.
These are very common; as one substance loses electrons the other substance
accepts them.
-- Electrochemical
process are redox reactions where energy is produced by a spontaneous reaction
which produces electricity, otherwise electrical current stimulates a chemical
reaction. In a redox reaction, an atom's oxidation state changes as a result
of an electron transfer.
-- A spontaneous
electrochemical reaction can be used to generate an electrical current,
in electrochemical cells. This is the basis of all batteries and fuel cells.
For example, gaseous oxygen (O2) and hydrogen (H2) can be combined in a
fuel cell to form water and energy (a combination of heat and electrical
energy, typically).
-- Conversely,
non-spontaneous electrochemical reactions can be driven forward by the
application of a current at sufficient voltage. The electrolysis of water
into gaseous oxygen and hydrogen is a typical example.
-- NADPH a
universal reductant used to reduce CO2 into sugars as well as for other
biosynthetic reductions.
-- The three-dimensional
tertiary structure of bacteriorhodopsin resembles that of vertebrate rhodopsins,
the pigments that sense light in the retina. Rhodopsins also contain retinal,
however the functions of rhodopsin and bacteriorhodopsin are different
and there is no homology of their amino acid sequences.
-- The electron
transfer reactions concentrate protons inside the membrane vesicle and
create an electric field across the photosynthetic membrane.
-- Protons
pass through the ATP-Synthase protein complex that transforms electrochemical
free energy into a type of chemical free energy known as phosphate group-transfer
potential (or a high-energy phosphate bond). The energy stored in ATP can
be transferred to another molecule by transferring the phosphate group.
-- The
oxidation- reduction midpoint potential (Em,7) of water is +0.82 V (pH
7). In photosystem II this reaction is driven by the oxidized reaction
center, P680+ (the midpoint potential of P680/P680+ is estimated to be
+1.2 V at pH 7)
--------------------------------------------------------
Life is right handed
Some molecules,
like sugars, come in both left handed and right handed versions. When made
in the laboratory from symmetric non-organic sources, you get roughly an
even mixture of right and left handed molecules, which are chemically identical.
Feed this sugar mixture to sugar eating bacteria and you find the bacteria
eat only half the sugar. The bacteria eat the right handed sugar
and leave the left handed sugar. Amazingly all life, everything
living on earth, at the molecular level is right handed.
The reason
bacteria can't eat left handed sugars is because many of the cells' complex
key molecules, proteins and especially enzyme proteins, which act as catalysts
greatly speeding biological operations, fold up in a way that reflects
their handedness. The simple picture of how enzymes work is 'lock and key'.
The enzyme orients the reacting molecule and keeps in position to react
because the reacting molecule fits within a nook (fold) of the enzyme.
So even though
life is based on chemistry and right and left handed sugars are chemically
identical, the lack of fit between left handed sugars and right
handed enzymes means that cells can't process left handed sugars.
Evolutionary consequences
As far as is known,
the choice of handedness when life begins is random. If this is right,
then clearly the fact that all life that has ever seen on earth
has the same handedness is strong evidence that life on earth probably
originated (or arrived !) just once, or more accurately, that it did not
independently originate (or arrive) many times.
------------------------------------------------------------------
--
ATP is the chemical equivalent of a loaded spring; the close packaging
of the three negatively charged phosphate groups ia an unstable, energy-storing
arrangement (like charges repel). The chemical “spring” tends to
“relax” from the loss of terminal phosphate. The cell taps this energy
source by using enzymes (kinases) to transfer phosphate groups from ATP
to other compounds, which are then said to be phosphorylated. Adding
the phosphate primes a molecule to undergo some kind of change that performs
work, and the molecule loses its phosphate group in the process.
-- ATP (Adenosine
triphosphate) It is the most important "molecular currency" of intracellular
energy transfer. (C10H16N5O13P3). It is produced as an energy source
during the processes of photosynthesis and cellular respiration and consumed
by many enzymes. This large release in energy makes the decomposition of
ATP in water extremely exergonic (-20.5 kJ / mole, with a change in free
energy of 3.4 kJ/mole). The overall process of oxidizing glucose to carbon
dioxide is known as cellular respiration and can produce up to 30 molecules
of ATP from a single molecule of glucose
-- The system
of ATP and water under standard conditions and concentrations is extremely
rich in chemical energy; the bond between the second and third phosphate
groups is loosely said to be particularly high in energy. Strictly speaking,
the bond itself is not high in energy (like all chemical bonds it requires
energy to break), but energy is produced when the bond is broken and water
is allowed to react with the two products. Thus, energy is produced from
the new bonds formed between ADP and water, and between phosphate and water.
-- The phosphate
ion is a polyatomic ion with the empirical formula PO4 and a molar mass
of 94.973 g/mol; it consists of one central phosphorus atom surrounded
by four identical oxygen atoms in a tetrahedral arrangement. It has a charge
of -3.
-- This gradient
is composed of both the pH gradient and the electrical gradient. The pH
gradient is a result of the H+ ion concentration difference. Together the
electrochemical gradient of protons is both a concentration and charge
difference and is often called the proton motive force (PMF).
-- In mitochondria
the PMF (proton motive force) is almost entirely made up of the electrical
component but in chloroplasts the PMF is made up mostly of the pH gradient.
In either case the PMF needs to be about 50 kJ/mol for the ATP synthase
to be able to make ATP.
** -- Mitchell (1978 Nobel prize for
Chemistry) realized that the movement of ions across an electrochemical
membrane potential could provide the energy needed to produce ATP. He knew
that living cells had a membrane potential; interior negative to the environment.
The movement of charged ions across a membrane is thus affected by the
electrical forces (the attraction of plus to minus charges). Their movement
is also affected by thermodynamic forces, the tendency of substances to
diffuse from regions of higher concentration. He went on to prove that
ATP synthesis was coupled to this electrochemical gradient.
-- Mitchell's
theory was confirmed by the discovery of ATP synthase, a membrane-bound
protein that uses the potential energy of the electrochemical gradient
to make ATP. Mitchell's chemiosmotic theory turned out to be one of the
two seminal discoveries in biology in the 20th century (DNA being the other)
-- Hydrogen
ions (protons) will diffuse from an area of high proton concentration to
an area of lower proton concentration. Peter Mitchell proposed that an
electrochemical concentration gradient of protons across a membrane could
be harnessed to make ATP. He likened this process to osmosis, the diffusion
of water across a membrane, which is why it is called chemiosmosis.
-- ATP synthase
is the enzyme that makes ATP by chemiosmosis. It allows protons to pass
through the membrane using the kinetic energy to phosphorylate ADP
making ATP. The generation of ATP by chemiosmosis occurs in chloroplasts
and mitochondria as well as in some bacteria.
-- Equation
for diffusion of gas across a cell membrane (Ficks first law)
Rate of Diffusion = k (A/d) (pressure difference)
Note this is like a current equation (i = V/R = V/(resistivity x length/area),
so flow x pressure diff must be power
-- Osmosis
is the net movement of a solvent across a semipermeable membrane
from a region of high solvent potential to an area of low solvent potential.
(Note it is the solvent that moves not the solute!) It is a physical process
in which a solvent moves, without input of energy, across a semipermeable
membrane (permeable to the solvent, but not the solute) separating two
solutions of different concentrations. Osmosis releases energy,
and can be made to do work. In plant cells, water enters the cell until
the inside and outside water potential is equal.
-- Cellular respiration
is a process that describes the metabolic reactions and processes that
take place in a cell to obtain chemical energy from fuel molecules. Energy
is released by the oxidation of fuel molecules and is stored as "high-energy"
carriers. The reactions involved in respiration are catabolic reactions
in metabolism. Fuel molecules commonly used by cells in respiration include
glucose, xx, and a common oxidizing agent (electron acceptor) is molecular
oxygen (O2).
-- The energy
released in respiration is used to synthesize molecules that act as a chemical
storage of this energy. One of the most widely used compounds in a cell
is adenosine triphosphate (ATP). Most of the ATP produced by cellular respiration
is by oxidative phosphorylation?? Chemiosmotic hypothesis (1961)
--- The theory suggests essentially that most ATP synthesis in respiring
cells comes from the electrochemical gradient across the inner membranes
of mitochondria.
-- The
energy available in the electrons is used to pump protons from the matrix
across the inner mitochondrial membrane, storing energy in the form of
a transmembrane electrochemical gradient. The electrons and protons at
the last pump in the ETC are taken up by oxygen to form water.
-- Chemiosmosis
is the diffusion of ions across a membrane. More specifically, it relates
to the generation of ATP by the movement of hydrogen ions across a membrane.
It allows protons to pass through the membrane using the kinetic energy
to phosphorylate ADP making ATP. The generation of ATP by chemiosmosis
occurs in chloroplasts and mitochondria as well as in some bacteria.
-- (the proton
circulate in and out of the cell. In driven by the electrochemical potential
to make ATP and then some of the energy available is used to pump them
out again restoring the potential. And some protons inside are oxidized
to water). The energy available in the electrons (from metabolized glucose)
is used to pump protons from the matrix across the inner mitochondrial
membrane, storing energy in the form of a transmembrane electrochemical
gradient. The protons move back across the inner membrane through the enzyme
ATP synthase
-- ATP is made
by an enzyme called ATP synthase. The structure of this enzyme and its
underlying genetic code is remarkably similar in all known forms of life.
ATP synthase is powered by a transmembrane electrochemical potential gradient,
usually in the form of a proton gradient. The function of the electron
transport chain is to produce this gradient.
-- The potential
energy that an electron has is determined by its distance from the nucleus.
The more energy the electron contains, the further it will be from the
nucleus; an electron with low energy will be closer to the nucleus. Electrons
can move to a higher energy level by having added to it (sunlight and light
energy). When this electron moves back to its original position, the same
amount of energy that it took to move the electron is released.
-- Most carbohydrates
have the empirical formula C(H2O)n. Carbohydrates are composed of covalently
bonded atoms of carbon, hydrogen, and oxygen.
-- The basic unit of a carbohydrate is a monosaccharide or simple sugar.
Monosaccharides can be burned (oxidized) to yield carbon dioxide, water,
and energy. The principle source of energy for organisms is glucose ( C6H12O6).
Structurally a sugar consists of a carbon backbone of three or more carbon
atoms with either an aldehyde or carbonyl group on one carbon and hydroxyl
groups on each of the other carbons.
-- Electron
carriers Some compounds can accept and donate electrons readily,
and these are called electron carriers in organisms. NADP is an electron
carrier used in photosynthesis. These molecules readily give up 2 electrons
(oxidized) and gain two electrons (reduced). Along with the electrons the
molecules accept 2 hydrogens to offset the negative charge of the electrons.
-- Oxidation
Losing
electrons, hydrogen or the gaining of oxygen. An electron loses potential
energy when it shifts from a less eletronegative atom towards a more electronegative
one. A redox reaction that relocates electrons closer to oxygen realeases
chemical energy which can be put to work.
-- Changing
in Covalent Status Usually, organic molecules that have and abundance
of hydrogen are excellent fuels because their bonds are a source of electrons
with high potential energy. They also have the potential to
drop the energy when they move closer to oxygen. The important
point in aerobic respiration is the change in covalent status of electrons
as hydrogen is transferred to oxygen.This is what liberates the energy.
-- At
key steps in aerobic respiration, hydrogen atoms are stripped from glucose,
but they are not directly transferred to oxygen. They are passed
to a coenzyme called NAD+ (nicotinamide adenine dinucleotide) which
functions as the oxidizing agent. Enzymes called dehydrogenases remove
a pair of hydrogen atoms from the substrate. These enzymes deliver
two electrons along with one proton to NAD+, forming NADH. The other
proton is released as a hydrogen ion into the surrounding solution.
-- Electrons
lose very little potential energy when they are transferred by dehydrogenases
from glucose (organic molecules) to NAD+. Thus, each NADH molecules
formed during respiration represents stored energy that can be used to
make ATP when the electrons complete their journey from NADH to oxygen.
-- (Electrons
can lower their energy by pairing {canceling magnetic moment??} This explains
a lot of bonding and chemistry) --- Two atoms of hydrogen, each with one
proton and one orbiting electron, will join into a dumbbell-shaped hydrogen
molecule. With the two electrons now paired, the molecule has a lower energy
state than the two separate hydrogen atoms. For this reason molecules with
an odd number of electrons are highly unstable, likely to react with the
first molecule that passes by.
** -- Compared to the inner membranes
of mitochondria, which have a significantly higher membrane potential due
to charge separation, thylakoid membranes lack a charge gradient.
To compensate for this, the 10,000 fold proton concentration gradient
across the thylakoid membrane is much higher compared to a 10 fold gradient
across the inner membrane of mitochondria.
-----------------------------------------------------------------------------------
(This section on pn junction may be out of date)
pn junctions ---electric field and diffusion gradients
A pn junction
is formed in a semiconductor when the fixed dopant atoms in the crystal
change (over a very short distance) from acceptor (P type) to donar (N
type). The resultant pn junction has steep charge gradients of mobile positive
(holes) and negative (electrons) carriers across the junction and an intrinsic
voltage across the junction that opposes charge flow along the gradients.
This voltage across the junction is initially set up by a small amount
of positive charge (holes) from the P side diffusing to the N side, and
a small amount of negative charge (electrons) diffusing from the N side
to the P side. The holes and electrons that cross the junction annihilate
each other (electrons fall into holes) expose in a thin region (depletion
region) spanning the junction the underlying non-mobile charge in the crystal,
which is positive on the N side of the junction and negative on the P side.
The non-mobile
exposed charge forms an electric field that points from the N to the P
side. This electric field in the depletion region sweeps holes back to
the P side and electrons back to the N side, thus keeping the depletion
region free of mobile charge and (in effect) preventing charge from moving
along the diffusion gradients. Summarizing, across a pn junction there
exists two strong charge moving mechanisms, diffusion and an E field, that
are opposed, and if undisturbed quickly come into balance preventing any
net charge motion across the junction.
Small changes
to the junction field by an external voltage can induce large diffusion
of charge carriers across the junction. For example: current in a diode
increases exponentially if the voltage across the diode (P side positive
with respect to N) is increased even slightly above the what is known as
the diode forward voltage (typ 700 mv for a silicon diode). An analogy
(from a 50's Scientific American article) is a dam with an overflow spillway.
A slight lowering of the dam height (junction voltage) causes a large flow
of water (diffusing charges) across the dam.
Consider what
happens when light falls on a pn junction (solar cell). Electron/hole pairs
are generated by each photon of light and the charges are accelerated (in
opposite directions) by the voltage across the junction. The holes driven
by the electric field to the P side and the electrons to the N side. This
raises the potential of the P side with respect to the N side, so if wires
are attached to the P and N materials and resistor connected between them
current will flow from the P material through the resistor to the N material.
The power dissipated in the resistor has come from the energy of the photons
via the electric field across the junction.
I wonder ---
Does understanding the energy physics of pn solar cells help us understand
how photosynthesis works, how a plant is able via photosynthesis to capture
the energy of solar photons?
Here is a link to lectures on basic semiconductor physics
-- (discussion of why voltage is proportional to logarithm of charge densities)
http://inst.eecs.berkeley.edu/~ee130/sp03/lecture.html
Do cell membranes act like semiconductor pn junctions?
Question:
Do cell membranes act like pn junctions? --- Yes, somewhat. Cell
membranes usually have across them ion gradients and if the membrane pores
are 'open' (to diffusion), then an electric field soon forms that blocks
further ion (potassium, sodium, hydrogen) diffusion through the membrane.
In a common
case the resting potential across a membrane is set up by (a few)
potassium (K+) ions diffusing out of the cell (and negative ions diffusing
in). The tendency of K+ to flow down the diffusion gradient out of the
cell is balanced by the voltage across the membrane driving K+ into the
cell. The membrane is spoken of a have a membrane capacitance and
the voltage across it is due to the excess charges next to the membrane.
The diffusion channel of a membrane is modeled as a resistor with
a different ohms for each type of ion. Note the ion channel in the membrane
must be open for the voltage to appear. The resting membrane voltage generated
by potassium is -80 mv. Typical capacitance is 66 pf and typical resistance
is 33 Mohm (RC = about 2 msec). A neuron cell fires for 2 msec passing
a current of 3.3 na. (see 'membrane potential')
--------------------------------------------------------------------
ATP
ATP is to
the cell what hydrogen might someday be to cars. It is not really a fuel,
it's an energy transport molecule. It's used in the cell to power basic
functions like ion pumps and muscle cells use it to power a contraction,
but ATP is not transported through the body, it's made locally in each
cell. Energy is transported around the body via sugars, fats, etc. The
cell's mitochondria organelles take in these sugars, oxidized them, and
use the energy released to build up an energy store called, a 'proton motive
force', which is later used to make ATP.
The energy
from burned sugar, in the form of an electron transport chain (similar
to photosynthesis!), runs proton pumps in the mitochondria that pump protons
out of the center region of mitochondria into a small inter-membrane space.
This proton pumping drives the inner mitochondria membrane voltage negative
(-190 mv), storing energy in the membrane capacitor, and some energy is
also stored in a small H+ gradient. When ATP needs to be made, the 'proton
motive force', meaning the membrane voltage and proton gradient, cause
a flow of protons through the membrane into the center of the mitochondria.
The protons flow in through the ATP synthase machine, which is a (9 nm
dia) 'turbine', causing it to actually spin (up to 12,000 RPM) and with
each 120 degree rotation an ADP is recombined with a Pi to make an ATP.
The ATP then (apparently) diffuses out of the mitochondria into other parts
of the cell to do work.
-- If an enzyme
is going to "push" two molecules together until they bond covalently, the
energy to do that has to come from somewhere. That "somewhere" is frequently
provided by a molecule of ATP. The enzyme, DNA polymerase, not only
has binding sites for the molecules it is going to join together, but another
binding site for a molecule of ATP. Once the enzyme has bound all the players
in the reaction, then the molecule of ATP is partially broken down; one
covalent bond in the ATP is broken.
Breaking that
covalent bond releases the energy that was put into it during its
formation. The energy released when this particular bond in the ATP is
broken is "caught" and harvested by the enzyme. The energy causes the enzyme
to change shape in a small way, so that the OTHER things it is holding
onto are forced together and covalently bonded together. SO, one covalent
bond is broken in ATP, and a new covalent bond is formed in the other molecules
the enzyme is binding. All that is left is for the enzyme to release everything
and start all over.
-- A critically
important macromolecule—arguably “second in importance only to DNA”—is
ATP.
-- As far as
known, all organisms from the simplest bacteria to humans use ATP as their
primary energy currency.
-- Energy
is usually liberated from the ATP molecule to do work in the cell by a
reaction that removes one of the phosphate-oxygen groups, leaving adenosine
diphosphate (ADP)
-- The ATP
is used for many cell functions including transport work moving substances
across cell membranes. It is also used for mechanical work, supplying the
energy needed for muscle contraction. It supplies energy not only to heart
muscle (for blood circulation) and skeletal muscle (such as for gross body
movement), but also to the chromosomes and flagella to enable them to carry
out their many functions.
-- A major
role of ATP is in chemical work, supplying the needed energy to synthesize
the multi-thousands of types of macromolecules that the cell needs to exist.
-- The ATP
synthase revolving door resembles a molecular water wheel that harnesses
the flow of hydrogen ions in order to build ATP molecules. Each revolution
of the wheel makes 3 ATP and requires the energy of about nine hydrogen
ions (more likely 12 H+ ions since latest thinking is H+/ATP = 4 or maybe
4.47) returning into the mitochondrial inner chamber (Goodsell, 1996, p.74).
Located on the ATP synthase are three active sites, each of which converts
ADP to ATP with every turn of the wheel. Under maximum conditions, the
ATP synthase wheel turns at a rate of up to 200 revolutions (12,000 RPM
per second, producing 600 ATPs during that second.
-- The
chloroplasts first convert the solar energy into ATP stored energy, which
is then used to manufacture storage carbohydrates which can be converted
back into ATP when energy is needed.
-- The ATP
molecule is composed of three components. At the centre is a sugar molecule,
ribose (the same sugar that forms the basis of RNA). Attached to one side
of this is a base (a group consisting of linked rings of carbon and nitrogen
atoms); in this case the base is adenine. The other side of the sugar is
attached to a string of phosphate groups. These phosphates are the key
to the activity of ATP.
-- Modern
fertilizers often contain phosphorus compounds that have been extracted
from animal bones. These compounds are used by plants to make ATP. We then
eat the plants, metabolise their phosphorus, and produce our own ATP.
--- ATP is
made by the enzyme ATP synthase. A Nobel prize for work on ATP synthase
was given only recently (1997).
-- Under certain
conditions, protons can re-enter the mitochondrial matrix without contributing
to ATP synthesis. This process is known as proton leak or mitochondrial
uncoupling and is due to the facilitated diffusion of protons into the
matrix. This process results in the unharnessed potential energy of the
proton electrochemical gradient being released as heat. (This heat can
be desired if the body needs to be warmed.)
----------------------------------
Fundamentals of life (1/30/15)
Here's an interesting speculation of the minimum set of characteristics
that are shared by every living thing on earth. The author, a biology professor,
calls it a list of 'minimum characteristics of the last common ancestor
of all life' from the book "Beginnings of Cellular Life" (by Prof Harold
Morowitz, 1992, Yale University Press).
All life
is cellular.
All
living things are from 50 to over 90% water, which is the source of protons,
hydrogen and oxygen in photosynthesis and the solvent of biomolecules.
The
major elements of covalently bound biomolecules are carbon, hydrogen, nitrogen,
oxygen, phosphorus and sulfur.
There
is a universal set of small molecules: (i.e. sugars, amino acids, nucleotides,
fatty acids, phospholipids, vitamins and coenzymes.)
The
principle macromolecules are proteins, lipids, carbohydrates and nucleic
acids.
There
is a universal type of membrane structure (i.e. the lipid bilayer).
The
flow of energy in living things involves formation and hydrolysis of phosphate
bonds, usually ATP.
The
metabolic reactions of any living species is a subset of a universal network
of intermediary metabolism (i.e. glycolysis; the Krebs cycle, the electron
transport chain)
Every
replicating cell has a genome made of DNA that stores the genetic information
of the cell which is read out in sequences of RNA and translated into protein.
All
growing cells have ribosomes, which are the sites of protein synthesis.
All
living things translate information from nucleotide language through specific
activating enzymes and transfer RNAs.
All
replicating biological systems give rise to altered phenotype due to mutated
genotypes.
Reactions
that proceed at appreciable rates in all living cells are catalyzed by
enzymes.






